Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Yokoya, Akinari; Fujii, Kentaro; Ushigome, Takeshi; Shikazono, Naoya; Urushibara, Ayumi; Watanabe, Ritsuko
Radiation Protection Dosimetry, 122(1-4), p.86 - 88, 2006/12
Times Cited Count:11 Percentile:60.11(Environmental Sciences)We have studied yields of DNA damages induced by soft X-rays obtained from a conventional soft X-ray machine in a LET region between  -rays and ultrasoft X-rays. Practically soft X-rays with a broad energy spectrum emitted from a target of heavy metal, such as tungsten, have been widely used not only for radiobiological experiments but also for medical application such as mammography. Radiation weighting factors for these soft X-rays have been assigned to be 1 by ICRP. However, the fraction of a large number of low energy photons in the spectrum (below several tens keV) provided by bremsstrahlung is expected to be more effective for DNA damage induction than
-rays and ultrasoft X-rays. Practically soft X-rays with a broad energy spectrum emitted from a target of heavy metal, such as tungsten, have been widely used not only for radiobiological experiments but also for medical application such as mammography. Radiation weighting factors for these soft X-rays have been assigned to be 1 by ICRP. However, the fraction of a large number of low energy photons in the spectrum (below several tens keV) provided by bremsstrahlung is expected to be more effective for DNA damage induction than  -rays since low energy photo- and Auger electrons predominantly ejected in consequence of a photoelectric effect can produce dense clusters of ionization/excitation on DNA molecules. We have examined the yield of DNA strand breaks induced by white soft X-rays (150 kVp, tungsten target). Yields of base lesions revealed by base excision repair enzymes will be also presented.
-rays since low energy photo- and Auger electrons predominantly ejected in consequence of a photoelectric effect can produce dense clusters of ionization/excitation on DNA molecules. We have examined the yield of DNA strand breaks induced by white soft X-rays (150 kVp, tungsten target). Yields of base lesions revealed by base excision repair enzymes will be also presented.
Pinak, M.
Modern Methods for Theoretical Physical Chemistry of Biopolymers, p.191 - 210, 2006/00
Results of molecular dynamics (MD) studies of several lesions on DNA and their respective repair enzymes are presented. Main focus is to describe structural and energy changes in DNA molecule with the respect to proper recognition of the lesion by respective repair enzyme. Pyrimidine and purine lesions were studied using MD simulation code Amber and respective force field modified for each lesion. The significant structural changes as breaking of hydrogen bonds network opening and bending the DNA double helix were observed. This collapsing of the double helical structure around the lesion is considered to facilitate docking of the repair enzyme into the DNA. Specific values of electrostatic interaction energy that enables repair enzyme to discriminate lesion from non-damaged site were also detected at several lesion sites.
Yokoya, Akinari; Shikazono, Naoya; Urushibara, Ayumi; Fujii, Kentaro; Akamatsu, Ken; Watanabe, Ritsuko
Hoshasen Seibutsu Kenkyu, 40(2), p.168 - 184, 2005/06
Ionizing radiation causes modifications in a DNA molecule depending on the characteristic tack-structure in which two or more isolated lesions arise in a few nm scale (1 or 2 helical turn of DNA), known as "clustered DNA damage". These clustered DNA damages could be distinct from those by reactive oxygen species (ROS) endogenously induced on their severity of induction of biological effects such as mutation. However, the studies on the nature and repair mechanism of clustered DNA damage have still been behind because of the technical difficulties on determination of the chemical structure and yield. This article reviews some experimental evidences of the clustered DNA damages in this research field, as well as our recent progress on the studies on the clustered DNA damages using both molecular biological techniques and synchrotron spectroscopic method.
Schyman, P.*; Danielsson, J.*; Pinak, M.; Laaksonen, A.*
Journal of Physical Chemistry A, 109(8), p.1713 - 1719, 2005/02
Times Cited Count:39 Percentile:77.29(Chemistry, Physical)We have examined the role of the catalytic lysine (Lys 249) in breaking the glycosidic bond of 8-oxoguanine in the enzyme human 8-oxoguanine DNA glycosylase. It has been assumed that this lysine acts as a nucleophile in a S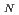 2 type of reaction after being activated through a donation of a proton to a strictly conserved aspartate. We use hybrid density functional theory to characterize both associative and dissociative pathways. We find that the smallest energetical barrier involves a S
2 type of reaction after being activated through a donation of a proton to a strictly conserved aspartate. We use hybrid density functional theory to characterize both associative and dissociative pathways. We find that the smallest energetical barrier involves a S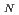 1 type of mechanism where the lysine electrostatically stabilizes the dissociating base and then donates a proton with a very small barrier and then finally attacks the sugar ring to create the covalently bounded protein-DNA intermediate complex. Reported findings give further support to the assumption that a dissociative mechanism may be the preferred mode of action for this type of enzymes.
1 type of mechanism where the lysine electrostatically stabilizes the dissociating base and then donates a proton with a very small barrier and then finally attacks the sugar ring to create the covalently bounded protein-DNA intermediate complex. Reported findings give further support to the assumption that a dissociative mechanism may be the preferred mode of action for this type of enzymes.
Pinak, M.; O'Neill, P.*; Fujimoto, Hirofumi; Nemoto, Toshiyuki*
AIP Conference Proceedings 708, p.310 - 313, 2004/05
The multiple nanosecond molecular dynamics simulations of DNA mutagenic oxidative lesion - 7,8-dihydro-8-oxoguanine (8-oxoG), complexed with the repair enzyme - human oxoguanine glycosylase 1 (hOGG1) in a physiological aqueous environment, were performed in order to describe the structural and energy changes in DNA and the dynamical process of DNA-enzyme complex formation. In complex the N-terminus of arginine 324 was found located close to the phosphodiester bond of the nucleotide with 8-oxoG enabling chemical reaction(s) between the amino acid and the lesion. The recognition of lesion on DNA, its recognition by repair enzyme and the formation of stable DNA-enzyme complex are necessary conditions for the onset of the successful enzymatic repair process.
Pinak, M.
Central European Journal of Physics, 1(1), p.179 - 190, 2003/01
The molecular dynamics (MD) studies of several radiation originated lesions on the DNA molecules are presented with the respect to the proper recognition of the lesion by the respective repair enzyme. Several pyrimidine and purine lesions were subjected to the MD simulations for several hundreds picoseconds using MD simulation code AMBER 5.0 and respective force field modified for the lesion. In all cases the significant structural changes in the DNA double helical structure were observed. These changes were related to the overall collapsing double helical structure around the lesion and are supposed to facilitate the docking of the repair enzyme into the DNA in formation of DNA-enzyme complex. In addition to the structural changes, the specific values of electrostatic interaction energy were found at several lesion sites.
 -radiation
-radiationYokoya, Akinari; Cunniffe, S. M. T.*; Stevens, D. L.*; O'Neill, P.*
Journal of Physical Chemistry B, 107(3), p.832 - 837, 2003/01
Times Cited Count:26 Percentile:56.36(Chemistry, Physical)no abstracts in English